Summer of 2002
was the first time I was gainfully employed as an information science
student. I was able to use my nursing background on a wonderful project
at the University
of Washington Health Sciences Library called PrimeAnswers
(PA). The Deputy Director of the HSL, Deborah Ketchell, and her website
team had created an electronic portal to evidence-based medical information
for University of Washington Primary Care Physicians.
The PA team includes
two medical librarians and one computer specialist. Student assistants
complete the team for both library and technology needs. In spring 2002,
the team was looking for an indexing student assistant. When I saw the
request through the iSchool email, I applied for the position. Although
I did not have one of the qualifications for which they were looking,
"experience in indexing semantic relations," I did have another,
"background in health-related terminology." I secured the
job and then applied for Directed Field Work (DFW) credits. It was a
backwards approach, but the position certainly met DFW criteria and
was a working example of melding information science, medicine, and
technology.
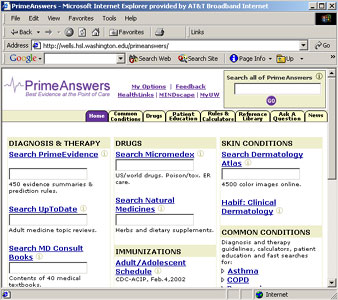
PrimeAnswers supports
the current model of evidence-based medicine. Physicians collect clinical
evidence through the physical exam, patient history and diagnostic tests.
In confirming the diagnosis, physicians also review external evidence
or randomized trials. Searching the literature for systematic research
is very difficult because of the sheer volume of current clinical literature.
The PrimeAnswers Web site was designed to make research evidence available
at the physician's fingertips, or "the best evidence at the point
of care."
The PA website
includes direct links to physician-preferred medical information sites
such as a drug information site (Micromedex) and medical question and
answer sites (UpToDate and MD Consult.) It also provides direct access
to articles from evidence-based medical journals on pertinent medical
topics or review articles on evidence-based studies. Another category
on the website is Common Conditions, a collection of frequently-encountered
conditions with brief but pertinent information about them.
When I began the
job in April of 2002, the website had been online for less than a year.
Many projects were still to be completed, so I got to see projects develop.
One of the most interesting projects was the creation of a controlled
vocabulary and thesaurus. Up to this point, searches on the website
had been carried out by keyword searching. I watched the process of
deciding whether to choose a commercial vendor environment for the thesaurus
or build our own. The resulting decision to build our own was based
on the expense of commercial products and the ability of the team computer
specialist to build a database "from scratch."
I began to see
what the terms Semantic Network, Modifiers, Categories, Controlled Terms,
and Preferred Terms meant when used in a medical thesaurus environment.
After the electronic data entry forms were created, the librarian team
members populated the thesaurus with pertinent terms from the database.
The stabilizing factor for placing terms in the thesaurus was connecting
each term to its counterpart term in the nationally-recognized medical
vocabulary called Unified Medical Language System (UMLS). Each term
was connected to a UMLS code number. Although medical terminology should
be standard, there are many preferences and differences nationwide and
worldwide. When the UMLS term did not match our purposes, the team could
apply a preferred term.
One of the most
interesting moments in website and thesaurus development came when the
librarians realized the fledgling thesaurus was not functioning the
way it was intended. A search for a certain term had to retrieve the
designated preferred term, either the UMLS term or the PA preferred
term. For example, a search for MI should return the term Myocardial
Infarction. Instead, synonymous terms were randomly returned. In order
to build this feature correctly, the head of the project had to be able
to speak both librarian and computer languages. This was, luckily, the
case.
My work on the
PA website involved indexing documents, mapping some of the more confusing
indexing systems on the database, and learning to make changes to the
public site by manipulating the administration tools "from the
backend."
But what I really
learned was how a project like this meets the definition of Bio-Informatics,
the newest melding of medicine and technology. It was great to see how
a librarian had created the fusion of the two disciplines.